General workflow demonstration¶
Setup and configuration¶
# # If you have JupyterLab, you will need to install the JupyterLab extension:
# # !jupyter labextension install @jupyter-widgets/jupyterlab-manager jupyter-leaflet
# On a normal notebook server, the extension needs only to be enabled
# !jupyter nbextension enable --py --sys-prefix ipyleaflet
import os
os.environ["USE_PYGEOS"] = "0" # force use Shapely with GeoPandas
import ipyleaflet
from birdy import WPSClient
from siphon.catalog import TDSCatalog
# we will use these coordinates later to center the map on Canada
canada_center_lat_lon = (52.4292, -93.2959)
PAVICS URL configuration¶
# Ouranos production server
pavics_url = "https://pavics.ouranos.ca"
finch_url = os.getenv("WPS_URL")
if not finch_url:
finch_url = f"{pavics_url}/twitcher/ows/proxy/finch/wps"
Getting user input from a map widget¶
Notes about ipyleaflet¶
ipyleaflet is a “A Jupyter / Leaflet bridge enabling interactive maps in the Jupyter notebook”
This means that the interactions with graphical objects on the map and in python are synchronized. The documentation is at: https://ipyleaflet.readthedocs.io
leaflet_map = ipyleaflet.Map(
center=canada_center_lat_lon,
basemap=ipyleaflet.basemaps.Stamen.Terrain,
zoom=4,
)
initial_marker_location = canada_center_lat_lon
marker = ipyleaflet.Marker(location=initial_marker_location, draggable=True)
leaflet_map.add_layer(marker)
leaflet_map
---------------------------------------------------------------------------
KeyError Traceback (most recent call last)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/xyzservices/lib.py:44, in Bunch.__getattr__(self, key)
43 try:
---> 44 return self.__getitem__(key)
45 except KeyError as err:
KeyError: 'Stamen'
The above exception was the direct cause of the following exception:
AttributeError Traceback (most recent call last)
Cell In[4], line 3
1 leaflet_map = ipyleaflet.Map(
2 center=canada_center_lat_lon,
----> 3 basemap=ipyleaflet.basemaps.Stamen.Terrain,
4 zoom=4,
5 )
7 initial_marker_location = canada_center_lat_lon
8 marker = ipyleaflet.Marker(location=initial_marker_location, draggable=True)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/xyzservices/lib.py:46, in Bunch.__getattr__(self, key)
44 return self.__getitem__(key)
45 except KeyError as err:
---> 46 raise AttributeError(key) from err
AttributeError: Stamen
If you move the marker on the map, it will update the marker.location variable. Also, if you update marker.location manually (marker.location = (45.44, -90.44)) it will also move on the map.
marker.location = (45.55, -72.44)
Helper function to get user input¶
import ipyleaflet
import ipywidgets as widgets
from IPython.display import display
def get_rectangle():
canada_center = (52.4292, -93.2959)
m = ipyleaflet.Map(
center=canada_center,
basemap=ipyleaflet.basemaps.Stamen.Terrain,
zoom=4,
)
# Create a new draw control
draw_control = ipyleaflet.DrawControl()
# disable some drawing inputs
draw_control.polyline = {}
draw_control.circlemarker = {}
draw_control.polygon = {}
draw_control.rectangle = {
"shapeOptions": {
"fillColor": "#4ae",
"color": "#4ae",
"fillOpacity": 0.3,
}
}
output = widgets.Output(layout={"border": "1px solid black"})
rectangle = {}
# set drawing callback
def callback(control, action, geo_json):
if action == "created":
# note: we can't close the map or remove it from the output
# from this callback. The map keeps the focus, and the
# jupyter keyboard input is messed up.
# So we set it very thin to make it disappear :)
m.layout = {"max_height": "0"}
with output:
print("*User selected 1 rectangle*")
rectangle.update(geo_json)
draw_control.on_draw(callback)
m.add_control(draw_control)
with output:
print("Select a rectangle:")
display(m)
display(output)
return rectangle
The user wants to select a rectangle on a map, and get a GeoJSON back¶
# NBVAL_IGNORE_OUTPUT
rectangle = get_rectangle()
# GeoJSON with custom style properties
if not len(rectangle):
# Use the default region of Greater Montreal Area
rectangle = {
"type": "Feature",
"properties": {
"style": {
"stroke": True,
"color": "#4ae",
"weight": 4,
"opacity": 0.5,
"fill": True,
"fillColor": "#4ae",
"fillOpacity": 0.3,
"clickable": True,
}
},
"geometry": {
"type": "Polygon",
"coordinates": [
[
[-74.511948, 45.202296],
[-74.511948, 45.934852],
[-72.978537, 45.934852],
[-72.978537, 45.202296],
[-74.511948, 45.202296],
]
],
},
}
rectangle
{'type': 'Feature',
'properties': {'style': {'stroke': True,
'color': '#4ae',
'weight': 4,
'opacity': 0.5,
'fill': True,
'fillColor': '#4ae',
'fillOpacity': 0.3,
'clickable': True}},
'geometry': {'type': 'Polygon',
'coordinates': [[[-74.511948, 45.202296],
[-74.511948, 45.934852],
[-72.978537, 45.934852],
[-72.978537, 45.202296],
[-74.511948, 45.202296]]]}}
Get the maximum and minimum bounds¶
import geopandas as gpd
rect = gpd.GeoDataFrame.from_features([rectangle])
bounds = rect.bounds
bounds
| minx | miny | maxx | maxy | |
|---|---|---|---|---|
| 0 | -74.511948 | 45.202296 | -72.978537 | 45.934852 |
Calling wps processes¶
For this example, we will subset a dataset with the user-selected bounds, and launch a heat wave frequency analysis on it.
finch = WPSClient(finch_url, progress=False)
help(finch.subset_bbox)
Help on method subset_bbox in module birdy.client.base:
subset_bbox(resource=None, lon0=0.0, lon1=360.0, lat0=-90.0, lat1=90.0, start_date=None, end_date=None, variable=None) method of birdy.client.base.WPSClient instance
Return the data for which grid cells intersect the bounding box for each input dataset as well as the time range selected.
Parameters
----------
resource : ComplexData:mimetype:`application/x-netcdf`, :mimetype:`application/x-ogc-dods`
NetCDF files, can be OPEnDAP urls.
lon0 : float
Minimum longitude.
lon1 : float
Maximum longitude.
lat0 : float
Minimum latitude.
lat1 : float
Maximum latitude.
start_date : string
Initial date for temporal subsetting. Can be expressed as year (%Y), year-month (%Y-%m) or year-month-day(%Y-%m-%d). Defaults to first day in file.
end_date : string
Final date for temporal subsetting. Can be expressed as year (%Y), year-month (%Y-%m) or year-month-day(%Y-%m-%d). Defaults to last day in file.
variable : string
Name of the variable in the NetCDF file.
Returns
-------
output : ComplexData:mimetype:`application/x-netcdf`
netCDF output
ref : ComplexData:mimetype:`application/metalink+xml; version=4.0`
Metalink file storing all references to output files.
Subset tasmin and tasmax datasets¶
# gather data from the PAVICS data catalogue
catalog = "https://pavics.ouranos.ca/twitcher/ows/proxy/thredds/catalog/datasets/gridded_obs/catalog.xml" # TEST_USE_PROD_DATA
cat = TDSCatalog(catalog)
data = cat.datasets[0].access_urls["OPENDAP"]
data
'https://pavics.ouranos.ca/twitcher/ows/proxy/thredds/dodsC/datasets/gridded_obs/nrcan_v2.ncml'
# NBVAL_IGNORE_OUTPUT
# If we want a preview of the data, here's what it looks like:
import xarray as xr
ds = xr.open_dataset(data, chunks="auto")
ds
<xarray.Dataset>
Dimensions: (lat: 510, lon: 1068, time: 24837)
Coordinates:
* lat (lat) float32 83.46 83.38 83.29 83.21 ... 41.29 41.21 41.12 41.04
* lon (lon) float32 -141.0 -140.9 -140.8 -140.7 ... -52.21 -52.13 -52.04
* time (time) datetime64[ns] 1950-01-01 1950-01-02 ... 2017-12-31
Data variables:
tasmin (time, lat, lon) float32 dask.array<chunksize=(3375, 69, 144), meta=np.ndarray>
tasmax (time, lat, lon) float32 dask.array<chunksize=(3375, 69, 144), meta=np.ndarray>
pr (time, lat, lon) float32 dask.array<chunksize=(3375, 69, 144), meta=np.ndarray>
Attributes: (12/15)
Conventions: CF-1.5
title: NRCAN ANUSPLIN daily gridded dataset : version 2
history: Fri Jan 25 14:11:15 2019 : Convert from original fo...
institute_id: NRCAN
frequency: day
abstract: Gridded daily observational dataset produced by Nat...
... ...
dataset_id: NRCAN_anusplin_daily_v2
version: 2.0
license_type: permissive
license: https://open.canada.ca/en/open-government-licence-c...
attribution: The authors provide this data under the Environment...
citation: Natural Resources Canada ANUSPLIN interpolated hist...# NBVAL_IGNORE_OUTPUT
# wait for process to complete before running this cell (the process is async)
tasmin_subset = result_tasmin.get().output
tasmin_subset
'https://pavics.ouranos.ca/wpsoutputs/finch/d00d6fa4-b40a-11ee-acf8-0242ac130007/nrcan_v2_sub.ncml'
result_tasmax = finch.subset_bbox(
resource=data,
variable="tasmax",
lon0=lon0,
lon1=lon1,
lat0=lat0,
lat1=lat1,
start_date="1958-01-01",
end_date="1958-12-31",
)
# NBVAL_IGNORE_OUTPUT
# wait for process to complete before running this cell (the process is async)
tasmax_subset = result_tasmax.get().output
tasmax_subset
'https://pavics.ouranos.ca/wpsoutputs/finch/d1151ece-b40a-11ee-acf8-0242ac130007/nrcan_v2_sub.ncml'
Launch a heat wave frequency analysis with the subsetted datasets¶
help(finch.heat_wave_frequency)
Help on method heat_wave_frequency in module birdy.client.base:
heat_wave_frequency(tasmin=None, tasmax=None, thresh_tasmin='22.0 degC', thresh_tasmax='30 degC', window=3, freq='YS', check_missing='any', missing_options=None, cf_compliance='warn', data_validation='raise', variable=None, output_name=None, output_format='netcdf', csv_precision=None, output_formats=None) method of birdy.client.base.WPSClient instance
Number of heat waves over a given period. A heat wave is defined as an event where the minimum and maximum daily temperature both exceeds specific thresholds over a minimum number of days.
Parameters
----------
tasmin : ComplexData:mimetype:`application/x-netcdf`, :mimetype:`application/x-ogc-dods`
NetCDF Files or archive (tar/zip) containing netCDF files. Minimum surface temperature.
tasmax : ComplexData:mimetype:`application/x-netcdf`, :mimetype:`application/x-ogc-dods`
NetCDF Files or archive (tar/zip) containing netCDF files. Maximum surface temperature.
thresh_tasmin : string
The minimum temperature threshold needed to trigger a heatwave event.
thresh_tasmax : string
The maximum temperature threshold needed to trigger a heatwave event.
window : integer
Minimum number of days with temperatures above thresholds to qualify as a heatwave.
freq : {'YS', 'MS', 'QS-DEC', 'AS-JUL'}string
Resampling frequency.
check_missing : {'any', 'wmo', 'pct', 'at_least_n', 'skip', 'from_context'}string
Method used to determine which aggregations should be considered missing.
missing_options : ComplexData:mimetype:`application/json`
JSON representation of dictionary of missing method parameters.
cf_compliance : {'log', 'warn', 'raise'}string
Whether to log, warn or raise when inputs have non-CF-compliant attributes.
data_validation : {'log', 'warn', 'raise'}string
Whether to log, warn or raise when inputs fail data validation checks.
variable : string
Name of the variable in the NetCDF file.
output_name : string
Filename of the output (no extension).
output_format : {'netcdf', 'csv'}string
Choose in which format you want to receive the result. CSV actually means a zip file of two csv files.
csv_precision : integer
Only valid if output_format is CSV. If not set, all decimal places of a 64 bit floating precision number are printed. If negative, rounds before the decimal point.
Returns
-------
output : ComplexData:mimetype:`application/x-netcdf`, :mimetype:`application/zip`
The format depends on the 'output_format' input parameter.
output_log : ComplexData:mimetype:`text/plain`
Collected logs during process run.
ref : ComplexData:mimetype:`application/metalink+xml; version=4.0`
Metalink file storing all references to output files.
result = finch.heat_wave_frequency(
tasmin=tasmin_subset,
tasmax=tasmax_subset,
thresh_tasmin="14 degC",
thresh_tasmax="22 degC",
)
Get the output as a python object (xarray Dataset)
response = result.get(asobj=True)
Downloading to /tmp/tmpqet24mhs/out.nc.
ds = response.output
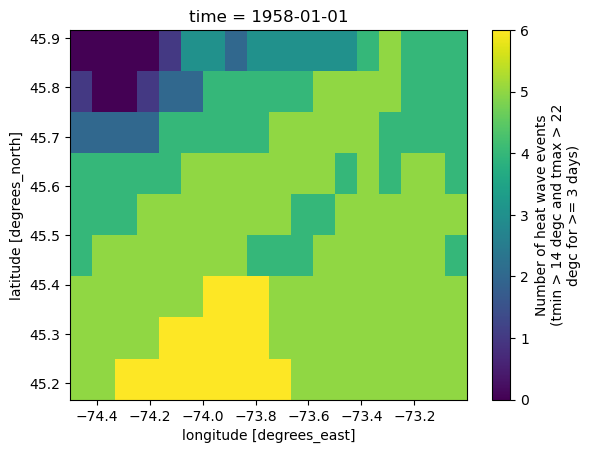
ds.heat_wave_frequency.plot()
<matplotlib.collections.QuadMesh at 0x7fccd44f0df0>
Streaming the data and showing on a map¶
The results are stored in a folder that is available to thredds, which provides a multiple services to access its datasets. In the case of large outputs, the user could view ths results of the analysis through an OPeNDAP service, so only the data to be shown is downloaded, and not the whole dataset.
Get the OPeNDAP url from the ‘wpsoutputs’¶
from urllib.parse import urlparse
output_url = result.get().output
print("output_url = ", output_url)
parsed = urlparse(output_url)
output_path = parsed.path.replace("wpsoutputs", "wps_outputs")
print("output_path = ", output_path)
output_thredds_url = (
f"https://{parsed.hostname}/twitcher/ows/proxy/thredds/dodsC/birdhouse{output_path}"
)
print("output_thredds_url = ", output_thredds_url)
output_url = https://pavics.ouranos.ca/wpsoutputs/finch/d2117e94-b40a-11ee-acf8-0242ac130007/out.nc
output_path = /wps_outputs/finch/d2117e94-b40a-11ee-acf8-0242ac130007/out.nc
output_thredds_url = https://pavics.ouranos.ca/twitcher/ows/proxy/thredds/dodsC/birdhouse/wps_outputs/finch/d2117e94-b40a-11ee-acf8-0242ac130007/out.nc
This time, we will be using hvplot to build our figure. This tool is a part of the holoviz libraries and adds an easy interface to xarray datasets. Since geoviews and cartopy are also installed, we can simply pass geo=True and a choice for tiles to turn the plot into a map. If multiple times were present in the dataset, a slider would appear, letting the user choose the slice. As we are using an OPeNDAP link, only the data that needs to be plotted (the current time slice) is downloaded.
# NBVAL_IGNORE_OUTPUT
import hvplot.xarray
dsremote = xr.open_dataset(output_thredds_url)
dsremote.hvplot.quadmesh(
"lon",
"lat",
"heat_wave_frequency",
geo=True,
alpha=0.8,
frame_height=540,
cmap="viridis",
tiles="CartoLight",
)
/opt/conda/envs/birdy/lib/python3.9/site-packages/geoviews/operation/__init__.py:14: HoloviewsDeprecationWarning: 'ResamplingOperation' is deprecated and will be removed in version 1.17, use 'ResampleOperation2D' instead.
from holoviews.operation.datashader import (