Analyzing the ClimEx Large Ensemble¶
ClimEx is a project investigating the influence of climate change on hydro-meteorological extremes. To understand the effect of natural variability on the occurrence of rare extreme events, ClimEx ran 50 independent simulations, increasing the sample size available.
Analyzing simulation outputs from large ensembles can get tedious due to the large number of individual netCDF files to handle. To simplify data access, we’ve created an aggregated view of daily precipitation and temperature for all 50 ClimEx realizations from 1955 to 2100.
The first step is to open the dataset, whose path can be found in the climex catalog. Although there are currently 36 250 individual netCDF files, there is only one link in the catalog.
import warnings
import numba
warnings.simplefilter("ignore", category=numba.core.errors.NumbaDeprecationWarning)
import logging
import intake
import xarray as xr
import xclim
from clisops.core.subset import subset_gridpoint
from dask.diagnostics import ProgressBar
from dask.distributed import Client, LocalCluster
from IPython.display import HTML, Markdown, clear_output
from xclim import ensembles as xens
# fmt: off
climex = "https://pavics.ouranos.ca/catalog/climex.json" # TEST_USE_PROD_DATA
cat = intake.open_esm_datastore(climex)
# fmt: on
cat.df.head()
| license_type | title | institution | driving_model_id | driving_experiment | type | processing | project_id | frequency | modeling_realm | variable_name | variable_long_name | path | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | permissive non-commercial | The ClimEx CRCM5 Large Ensemble | Ouranos Consortium on Regional Climatology and... | CCCma-CanESM2 | historical, rcp85 | RCM | raw | CLIMEX | day | atmos | ['tasmin', 'tasmax', 'tas', 'pr', 'prsn', 'rot... | ['Daily Minimum Near-Surface Temperature minim... | https://pavics.ouranos.ca/twitcher/ows/proxy/t... |
# Opening the link takes a while, because the server creates an aggregated view of 435,000 individual files.
url = cat.df.path[0]
ds = xr.open_dataset(url, chunks=dict(realization=2, time=30 * 3))
ds
<xarray.Dataset> Size: 4TB
Dimensions: (rlat: 280, rlon: 280, time: 52924, realization: 50)
Coordinates:
* rlat (rlat) float64 2kB -12.61 -12.51 -12.39 ... 17.85 17.96 18.07
* rlon (rlon) float64 2kB 2.695 2.805 2.915 ... 33.17 33.28 33.39
* time (time) object 423kB 1955-01-01 00:00:00 ... 2099-12-30 00:0...
* realization (realization) |S64 3kB b'historical-r1-r10i1p1' ... b'histo...
lat (rlat, rlon) float32 314kB dask.array<chunksize=(280, 280), meta=np.ndarray>
lon (rlat, rlon) float32 314kB dask.array<chunksize=(280, 280), meta=np.ndarray>
Data variables:
rotated_pole |S64 64B ...
tasmin (realization, time, rlat, rlon) float32 830GB dask.array<chunksize=(2, 90, 280, 280), meta=np.ndarray>
tasmax (realization, time, rlat, rlon) float32 830GB dask.array<chunksize=(2, 90, 280, 280), meta=np.ndarray>
tas (realization, time, rlat, rlon) float32 830GB dask.array<chunksize=(2, 90, 280, 280), meta=np.ndarray>
pr (realization, time, rlat, rlon) float32 830GB dask.array<chunksize=(2, 90, 280, 280), meta=np.ndarray>
prsn (realization, time, rlat, rlon) float32 830GB dask.array<chunksize=(2, 90, 280, 280), meta=np.ndarray>
Attributes: (12/30)
Conventions: CF-1.6
DODS.dimName: string1
DODS.strlen: 0
EXTRA_DIMENSION.bnds: 2
NCO: "4.5.2"
abstract: The ClimEx CRCM5 Large Ensemble of high-resolut...
... ...
project_id: CLIMEX
rcm_version_id: v3331
terms_of_use: http://www.climex-project.org/sites/default/fil...
title: The ClimEx CRCM5 Large Ensemble
type: RCM
EXTRA_DIMENSION.string1: 1The CLIMEX dataset currently stores 5 daily variables, simulated by CRCM5 driven by CanESM2 using the representative concentration pathway RCP8.5:
pr: mean daily precipitation fluxprsn: mean daily snowfall fluxtas: mean daily temperaturetasmin: minimum daily temperaturetasmax: maximum daily temperature
There variables are stored along spatial dimensions (rotated latitude and longitude), time (1955-2100) and realizations (50 members). These members are created by first running five members from CanESM2 from 1850 to 1950, then small perturbations are applied in 1950 to spawn 10 members from each original member, to yield a total of 50 global realizations. Each realization is then dynamically downscaled to 12 km resolution over Québec.

ds.realization[:5].data.tolist()
[b'historical-r1-r10i1p1',
b'historical-r1-r1i1p1',
b'historical-r1-r2i1p1',
b'historical-r1-r3i1p1',
b'historical-r1-r4i1p1']
Creating maps of ClimEx fields¶
The data is on a rotated pole grid, and the actual geographic latitudes and longitudes of grid centroids are stored in the variables with the same name. We can plot the data directly using the native rlat and rlon coordinates easily.
# NBVAL_IGNORE_OUTPUT
from matplotlib import pyplot as plt
with ProgressBar():
field = ds.tas.isel(time=0, realization=0).load()
field.plot()
[ ] | 0% Completed | 181.94 us
[ ] | 0% Completed | 101.69 ms
[ ] | 0% Completed | 202.52 ms
[ ] | 0% Completed | 303.31 ms
[ ] | 0% Completed | 404.06 ms
[ ] | 0% Completed | 504.78 ms
[ ] | 0% Completed | 605.72 ms
[ ] | 0% Completed | 706.44 ms
[ ] | 0% Completed | 807.17 ms
[ ] | 0% Completed | 907.87 ms
[ ] | 0% Completed | 1.01 s
[ ] | 0% Completed | 1.11 s
[ ] | 0% Completed | 1.21 s
[ ] | 0% Completed | 1.31 s
[#################### ] | 50% Completed | 1.41 s
[#################### ] | 50% Completed | 1.51 s
[#################### ] | 50% Completed | 1.61 s
[############################## ] | 75% Completed | 1.71 s
[############################## ] | 75% Completed | 1.81 s
[########################################] | 100% Completed | 1.92 s
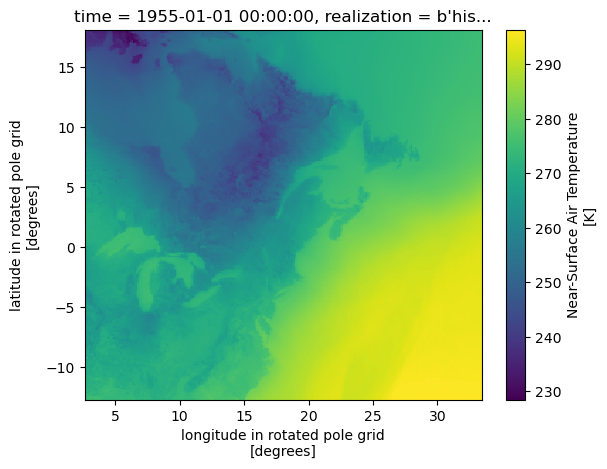
However, the axes are defined with respect to the rotated coordinates. To create a map with correct geographic coordinates, we could use the real longitudes and latitudes (plt.pcolormesh(ds.lon, ds.lat, field)), but a better option is to create axes with the same projection used by the model. That is, we set the map projection to RotatedPole, using the coordinate reference system defined in the rotated_pole variable. We can use a similar approach to project the model output on another projection.
# NBVAL_IGNORE_OUTPUT
import cartopy.crs as ccrs
with ProgressBar():
# The CRS for the rotated pole (data)
rotp = ccrs.RotatedPole(
pole_longitude=ds.rotated_pole.grid_north_pole_longitude,
pole_latitude=ds.rotated_pole.grid_north_pole_latitude,
)
# The CRS for your map projection (can be anything)
ortho = ccrs.Orthographic(central_longitude=-80, central_latitude=45)
fig = plt.figure(figsize=(8, 4))
# Plot data on the rotated pole projection directly
ax = plt.subplot(1, 2, 1, projection=rotp)
ax.coastlines()
ax.gridlines()
m = ax.pcolormesh(ds.rlon, ds.rlat, field)
plt.colorbar(
m, orientation="horizontal", label=field.long_name, fraction=0.046, pad=0.04
)
# Plot data on another projection, transforming it using the rotp crs.
ax = plt.subplot(1, 2, 2, projection=ortho)
ax.coastlines()
ax.gridlines()
m = ax.pcolormesh(
ds.rlon, ds.rlat, field, transform=rotp
) # Note the transform parameter
plt.colorbar(
m, orientation="horizontal", label=field.long_name, fraction=0.046, pad=0.04
)
plt.show()
ERROR 1: PROJ: proj_create_from_database: Open of /home/docs/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/share/proj failed
/home/docs/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/cartopy/io/__init__.py:241: DownloadWarning: Downloading: https://naturalearth.s3.amazonaws.com/50m_physical/ne_50m_coastline.zip
warnings.warn(f'Downloading: {url}', DownloadWarning)
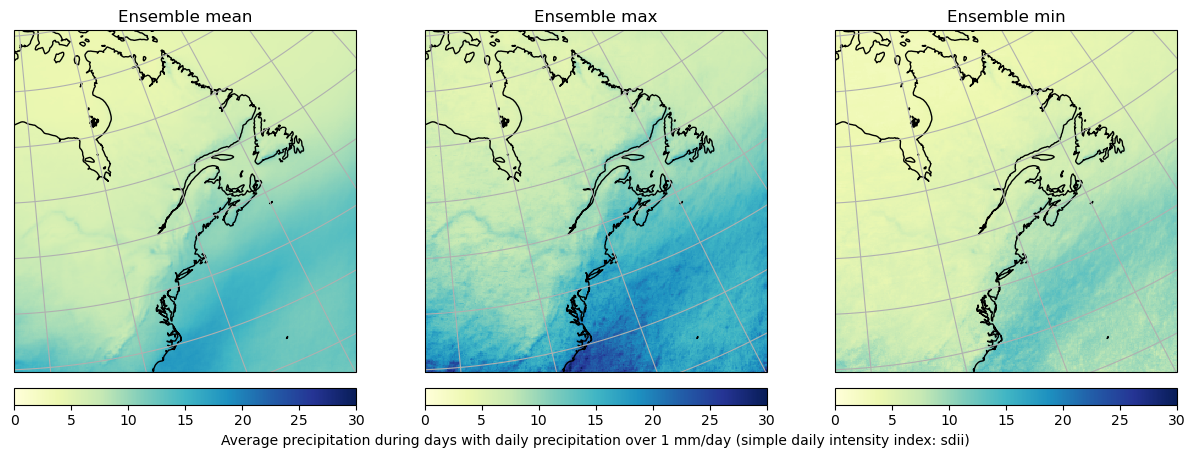
Computing climate indicators¶
Next we’ll compute a climate indicator as an example. We’ll compute the mean precipitation intensity for 50 members for the year 2000, then compute ensemble statistics between realizations. The amount of data that we’ll be requesting is however too big to go over the network in one request. To work around this, we need to first chunk the data in smaller dask.array portions by setting the chunks parameter when creating our dataset.
When selecting
xarraychunk sizes it is important to consider both the “on-disk” chunk sizes of the netcdf data as well as the type of calculation you are performing. For example the Climex data variables_ChunkSizesattributes indicate on-disk chunk size for dimstime, rlat, rlonof[365 50 50]1. Using multiples of these values is a reasonable way to ensure that in-memorydaskchunks line up with the way that the data is stored on disk.The next step is to think about the calculation we wish to perform. In this case we wish to analyse a single year of data for all realizations over the entire spatial domain. Using something like
chunks = dict(realization=1, time=365, rlat=50*3, rlon=50*3)seems reasonable as a start. The general goal is to find a balance between chunk-size and a number of chunks … too many small chunks to process will create a lot of overhead and slow the calculation down. Very large chunks will use a lot of system memory causing other issues or data timeouts from the PAVICS thredds server. In this specific case having a largetimechunk could result in reading more data than necessary seeing as we are only interested in a single year.Enabling chunking also has the effect of making further computations on these chunks lazy. The next cell is thus going to execute almost instantly, because no data is transferred, and no computation executed. Calculations are launched when the data is actually needed, as when plotting graphics, saving to file, or when requested explicitly by calling the compute, persist or load methods. Below we make use of a dask.distributed Client of worker processes to parallelize computations and improve performance.
1 In this case the `realization` chunk size is not included in the netcdf file attributes as this is a virtual dimension resulting from the NcML aggregation.
ds = xr.open_dataset(
url, chunks=dict(realization=1, time=365, rlat=50 * 3, rlon=50 * 3)
)
xclim.set_options(check_missing="pct", missing_options={"pct": {"tolerance": 0.05}})
sdii = xclim.atmos.daily_pr_intensity(pr=ds.pr.sel(time="2000"))
sdii
<xarray.DataArray 'sdii' (realization: 50, time: 1, rlat: 280, rlon: 280)> Size: 31MB
dask.array<where, shape=(50, 1, 280, 280), dtype=float64, chunksize=(1, 1, 150, 150), chunktype=numpy.ndarray>
Coordinates:
* rlat (rlat) float64 2kB -12.61 -12.51 -12.39 ... 17.85 17.96 18.07
* rlon (rlon) float64 2kB 2.695 2.805 2.915 ... 33.17 33.28 33.39
* realization (realization) |S64 3kB b'historical-r1-r10i1p1' ... b'histor...
lat (rlat, rlon) float32 314kB dask.array<chunksize=(150, 150), meta=np.ndarray>
lon (rlat, rlon) float32 314kB dask.array<chunksize=(150, 150), meta=np.ndarray>
* time (time) object 8B 2000-01-01 00:00:00
Attributes:
units: mm d-1
cell_methods: time: mean
history: [2024-04-16 18:26:26] sdii: SDII(pr=pr, thresh='1 mm/day'...
standard_name: lwe_thickness_of_precipitation_amount
long_name: Average precipitation during days with daily precipitatio...
description: Annual simple daily intensity index (sdii) or annual aver...# NBVAL_IGNORE_OUTPUT
# Create a local client with 6 workers processes.
dask_kwargs = dict(
n_workers=6,
threads_per_worker=6,
memory_limit="4GB",
local_directory="/notebook_dir/writable-workspace/tmp",
silence_logs=logging.ERROR,
)
with Client(**dask_kwargs) as client:
clear_output()
display(
HTML(
f'<div class="alert alert-info"> Consult the client <a href="{client.dashboard_link}" target="_blank"><strong><em><u>Dashboard</u></em></strong></a> or the <strong><em>Dask jupyter sidebar</em></strong> to follow progress ... </div>'
)
)
out = xens.ensemble_mean_std_max_min(sdii.to_dataset())
out = out.load()
clear_output()
INFO:distributed.http.proxy:To route to workers diagnostics web server please install jupyter-server-proxy: python -m pip install jupyter-server-proxy
INFO:distributed.scheduler:State start
INFO:distributed.scheduler: Scheduler at: tcp://127.0.0.1:37225
INFO:distributed.scheduler: dashboard at: http://127.0.0.1:8787/status
INFO:distributed.scheduler:Registering Worker plugin shuffle
---------------------------------------------------------------------------
PermissionError Traceback (most recent call last)
Cell In[7], line 12
1 # NBVAL_IGNORE_OUTPUT
2
3 # Create a local client with 6 workers processes.
4 dask_kwargs = dict(
5 n_workers=6,
6 threads_per_worker=6,
(...)
9 silence_logs=logging.ERROR,
10 )
---> 12 with Client(**dask_kwargs) as client:
13 clear_output()
14 display(
15 HTML(
16 f'<div class="alert alert-info"> Consult the client <a href="{client.dashboard_link}" target="_blank"><strong><em><u>Dashboard</u></em></strong></a> or the <strong><em>Dask jupyter sidebar</em></strong> to follow progress ... </div>'
17 )
18 )
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/client.py:1017, in Client.__init__(self, address, loop, timeout, set_as_default, scheduler_file, security, asynchronous, name, heartbeat_interval, serializers, deserializers, extensions, direct_to_workers, connection_limit, **kwargs)
1014 preload_argv = dask.config.get("distributed.client.preload-argv")
1015 self.preloads = preloading.process_preloads(self, preload, preload_argv)
-> 1017 self.start(timeout=timeout)
1018 Client._instances.add(self)
1020 from distributed.recreate_tasks import ReplayTaskClient
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/client.py:1219, in Client.start(self, **kwargs)
1217 self._started = asyncio.ensure_future(self._start(**kwargs))
1218 else:
-> 1219 sync(self.loop, self._start, **kwargs)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/utils.py:434, in sync(loop, func, callback_timeout, *args, **kwargs)
431 wait(10)
433 if error is not None:
--> 434 raise error
435 else:
436 return result
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/utils.py:408, in sync.<locals>.f()
406 awaitable = wait_for(awaitable, timeout)
407 future = asyncio.ensure_future(awaitable)
--> 408 result = yield future
409 except Exception as exception:
410 error = exception
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/tornado/gen.py:767, in Runner.run(self)
765 try:
766 try:
--> 767 value = future.result()
768 except Exception as e:
769 # Save the exception for later. It's important that
770 # gen.throw() not be called inside this try/except block
771 # because that makes sys.exc_info behave unexpectedly.
772 exc: Optional[Exception] = e
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/client.py:1284, in Client._start(self, timeout, **kwargs)
1281 elif self._start_arg is None:
1282 from distributed.deploy import LocalCluster
-> 1284 self.cluster = await LocalCluster(
1285 loop=self.loop,
1286 asynchronous=self._asynchronous,
1287 **self._startup_kwargs,
1288 )
1289 address = self.cluster.scheduler_address
1291 self._gather_semaphore = asyncio.Semaphore(5)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/deploy/spec.py:420, in SpecCluster.__await__.<locals>._()
418 await self._start()
419 await self.scheduler
--> 420 await self._correct_state()
421 if self.workers:
422 await asyncio.wait(
423 [
424 asyncio.create_task(_wrap_awaitable(w))
425 for w in self.workers.values()
426 ]
427 ) # maybe there are more
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/deploy/spec.py:374, in SpecCluster._correct_state_internal(self)
372 if isinstance(cls, str):
373 cls = import_term(cls)
--> 374 worker = cls(
375 getattr(self.scheduler, "contact_address", None)
376 or self.scheduler.address,
377 **opts,
378 )
379 self._created.add(worker)
380 workers.append(worker)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/nanny.py:200, in Nanny.__init__(self, scheduler_ip, scheduler_port, scheduler_file, worker_port, nthreads, loop, local_directory, services, name, memory_limit, reconnect, validate, quiet, resources, silence_logs, death_timeout, preload, preload_argv, preload_nanny, preload_nanny_argv, security, contact_address, listen_address, worker_class, env, interface, host, port, protocol, config, **worker_kwargs)
186 preload_nanny_argv = dask.config.get("distributed.nanny.preload-argv")
188 handlers = {
189 "instantiate": self.instantiate,
190 "kill": self.kill,
(...)
198 "plugin_remove": self.plugin_remove,
199 }
--> 200 super().__init__(
201 handlers=handlers,
202 connection_args=self.connection_args,
203 local_directory=local_directory,
204 needs_workdir=False,
205 )
207 self.preloads = preloading.process_preloads(
208 self, preload_nanny, preload_nanny_argv, file_dir=self.local_directory
209 )
211 self.death_timeout = parse_timedelta(death_timeout)
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/core.py:403, in Server.__init__(self, handlers, blocked_handlers, stream_handlers, connection_limit, deserialize, serializers, deserializers, connection_args, timeout, io_loop, local_directory, needs_workdir)
394 self._original_local_dir = local_directory
396 with warn_on_duration(
397 "1s",
398 "Creating scratch directories is taking a surprisingly long time. ({duration:.2f}s) "
(...)
401 "scratch data to a local disk.",
402 ):
--> 403 self._workspace = WorkSpace(local_directory)
405 if not needs_workdir: # eg. Nanny will not need a WorkDir
406 self._workdir = None
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/diskutils.py:128, in WorkSpace.__init__(self, base_dir)
127 def __init__(self, base_dir: str):
--> 128 self.base_dir = self._init_workspace(base_dir)
129 self._global_lock_path = os.path.join(self.base_dir, "global.lock")
130 self._purge_lock_path = os.path.join(self.base_dir, "purge.lock")
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/site-packages/distributed/diskutils.py:146, in WorkSpace._init_workspace(self, base_dir)
144 for try_dir in try_dirs:
145 try:
--> 146 os.makedirs(try_dir)
147 except FileExistsError:
148 try:
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/os.py:215, in makedirs(name, mode, exist_ok)
213 if head and tail and not path.exists(head):
214 try:
--> 215 makedirs(head, exist_ok=exist_ok)
216 except FileExistsError:
217 # Defeats race condition when another thread created the path
218 pass
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/os.py:215, in makedirs(name, mode, exist_ok)
213 if head and tail and not path.exists(head):
214 try:
--> 215 makedirs(head, exist_ok=exist_ok)
216 except FileExistsError:
217 # Defeats race condition when another thread created the path
218 pass
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/os.py:215, in makedirs(name, mode, exist_ok)
213 if head and tail and not path.exists(head):
214 try:
--> 215 makedirs(head, exist_ok=exist_ok)
216 except FileExistsError:
217 # Defeats race condition when another thread created the path
218 pass
File ~/checkouts/readthedocs.org/user_builds/pavics-sdi/conda/latest/lib/python3.10/os.py:225, in makedirs(name, mode, exist_ok)
223 return
224 try:
--> 225 mkdir(name, mode)
226 except OSError:
227 # Cannot rely on checking for EEXIST, since the operating system
228 # could give priority to other errors like EACCES or EROFS
229 if not exist_ok or not path.isdir(name):
PermissionError: [Errno 13] Permission denied: '/notebook_dir'
fig = plt.figure(figsize=(15, 5))
for ii, vv in enumerate([v for v in out.data_vars if "stdev" not in v]):
ax = plt.subplot(1, 3, ii + 1, projection=rotp)
ax.coastlines()
ax.gridlines()
m = ax.pcolormesh(
out.rlon, out.rlat, out[vv].isel(time=0), vmin=0, vmax=30, cmap="YlGnBu"
)
c = plt.colorbar(
m, orientation="horizontal", label=out[vv].long_name, fraction=0.046, pad=0.04
)
ax.set_title(f"Ensemble {vv.split('_')[-1]}")
if ii != 1:
c.set_label("")
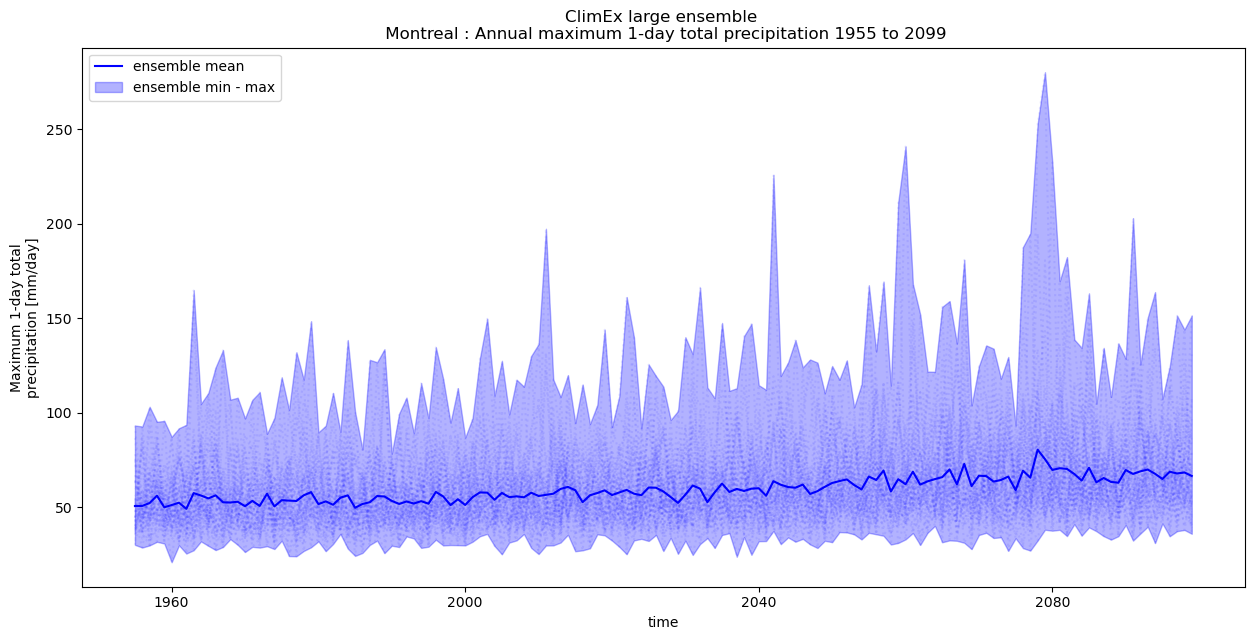
In the next example, we extract one grid point over Montréal in order to calculate the annual maximum 1-day precipitation for all years and ensemble members. In this case we would likely benefit by rethinking out chunk sizes.
Since we are only interested in a single grid-cell there is no real need to have relatively large spatial chunks in the
rlonandrlatdimensions read into memory to simply be immediately discarded. In this case we reduce the spatial chunk size.Similarly, since we are only looking at a single point we can likely get away with loading a large number of time steps into a single chunk.
# Subset over the Montreal gridpoint
ds = xr.open_dataset(url, chunks=dict(realization=1, time=365 * 50, rlon=25, rlat=25))
pt = subset_gridpoint(ds, lon=-73.69, lat=45.50)
print("Input dataset for Montreal :")
display(pt)
out = xclim.atmos.max_1day_precipitation_amount(pr=pt.pr, freq="YS")
print("Maximim 1-day precipitation `lazy` output ..")
out
Input dataset for Montreal :
<xarray.Dataset>
Dimensions: (time: 52924, realization: 50)
Coordinates:
rlat float64 0.365
rlon float64 16.12
* time (time) object 1955-01-01 00:00:00 ... 2099-12-30 00:00:00
* realization (realization) |S64 b'historical-r1-r10i1p1' ... b'historica...
lat float32 45.45
lon float32 -73.7
Data variables:
rotated_pole |S64 b''
tasmin (realization, time) float32 dask.array<chunksize=(1, 18250), meta=np.ndarray>
tasmax (realization, time) float32 dask.array<chunksize=(1, 18250), meta=np.ndarray>
tas (realization, time) float32 dask.array<chunksize=(1, 18250), meta=np.ndarray>
pr (realization, time) float32 dask.array<chunksize=(1, 18250), meta=np.ndarray>
prsn (realization, time) float32 dask.array<chunksize=(1, 18250), meta=np.ndarray>
Attributes: (12/30)
Conventions: CF-1.6
DODS.dimName: string1
DODS.strlen: 0
EXTRA_DIMENSION.bnds: 2
NCO: "4.5.2"
abstract: The ClimEx CRCM5 Large Ensemble of high-resolut...
... ...
project_id: CLIMEX
rcm_version_id: v3331
terms_of_use: http://www.climex-project.org/sites/default/fil...
title: The ClimEx CRCM5 Large Ensemble
type: RCM
EXTRA_DIMENSION.string1: 1Maximim 1-day precipitation `lazy` output ..
<xarray.DataArray 'rx1day' (realization: 50, time: 145)>
dask.array<where, shape=(50, 145), dtype=float32, chunksize=(1, 50), chunktype=numpy.ndarray>
Coordinates:
rlat float64 0.365
rlon float64 16.12
* realization (realization) |S64 b'historical-r1-r10i1p1' ... b'historical...
lat float32 45.45
lon float32 -73.7
* time (time) object 1955-01-01 00:00:00 ... 2099-01-01 00:00:00
Attributes:
units: mm/day
cell_methods: time: mean time: maximum over days
history: [2023-05-24 17:44:17] rx1day: RX1DAY(pr=pr, freq='YS') wi...
standard_name: lwe_thickness_of_precipitation_amount
long_name: Maximum 1-day total precipitation
description: Annual maximum 1-day total precipitation# NBVAL_IGNORE_OUTPUT
# Compute the annual max 1-day precipitation
dask_kwargs = dict(
n_workers=6,
threads_per_worker=6,
memory_limit="4GB",
local_directory="/notebook_dir/writable-workspace/tmp",
silence_logs=logging.ERROR,
)
with Client(**dask_kwargs) as client:
# dashboard available at client.dashboard_link
clear_output()
display(
HTML(
f'<div class="alert alert-info"> Consult the client <a href="{client.dashboard_link}" target="_blank"><strong><em><u>Dashboard</u></em></strong></a> or the <strong><em>Dask jupyter sidebar</em></strong> to follow progress ... </div>'
)
)
out = out.load()
clear_output()
out_ens = xens.ensemble_mean_std_max_min(out.to_dataset())
fig = plt.figure(figsize=(15, 7))
ax = plt.subplot(1, 1, 1)
h1 = out.plot.line(ax=ax, x="time", color="b", linestyle=":", alpha=0.075, label=None)
h2 = plt.fill_between(
x=out_ens.time.values,
y1=out_ens.rx1day_max,
y2=out_ens.rx1day_min,
color="b",
alpha=0.3,
label="ensemble min - max",
)
h3 = plt.plot(
out_ens.time.values, out_ens.rx1day_mean, color="b", label="ensemble mean"
)
handles, labels = ax.get_legend_handles_labels()
leg = ax.legend(handles[::-1], labels[::-1])
tit = plt.title(
f"ClimEx large ensemble \n Montreal : {out.description} {out_ens.time.dt.year[0].values} to {out_ens.time.dt.year[-1].values}"
)